So in my last post I promised to do a walk-through of how I made a contour plot of the function z = 6*x*y - 5*x2 using MATLAB. Here's a copy of that plot:
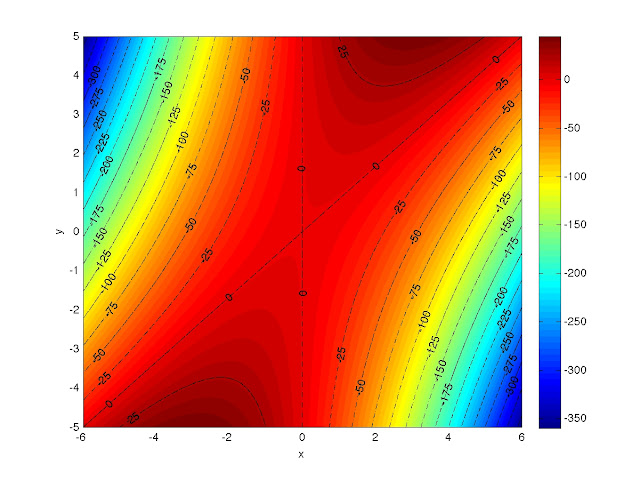 |
| Contour plot of z = 6*x*y - 5*x2. |
This will provide some illustration of some of the more abstract commands I mentioned in the last post, so let's get started. Remember: the semicolon character ( ; ) at the end of an expression will suppress the output from that line. If you accidentally do this for a plot command, it'll still show up though.
If you've been playing around in MATLAB already and you want to start from an empty workspace, type in the command
clear. You should notice that any workspace variables you had have magically vanished. If you want to clear out a specific quantity, type "clear quantity." Also, if you're following along and trying this yourself, realize that MATLAB is pretty forgiving with whitespace inside an expression, an argument to a command, or inside colon notation, so sometimes I add extra space to make things readable, sometimes I don't.
The first thing we need to do is make the coordinate grid. When I did this step for the previous post, I used colon notation:
X = -6:0.01:6;
Y = -5:0.01:5;
which produced two arrays: X, which is a 1201-element row vector from -6 to 6 in increments of 0.01; and Y, which is a 1001-element row vector from -5 to 5 in increments of 0.01. I know I haven't mentioned it yet, so let me say that if you don't give a middle argument in an expression that uses colon notation, it'll automatically increment by 1 between the two arguments. If the two bounding values aren't an integral multiple of the increment apart, the resulting array will not exceed the upper bound.
Since I know how many elements are in each vector, I can easily produce the same arrays using
linspace:
X = linspace(-6,6,1201);
Y = linspace(-5,5,1001);
Just for fun, I made Y with colon notation and then Ybar with linspace and then calculated the element-by-element difference between the two. The absolute values of the differences between corresponding elements ranged from 0 to about 9 x 10
-16.
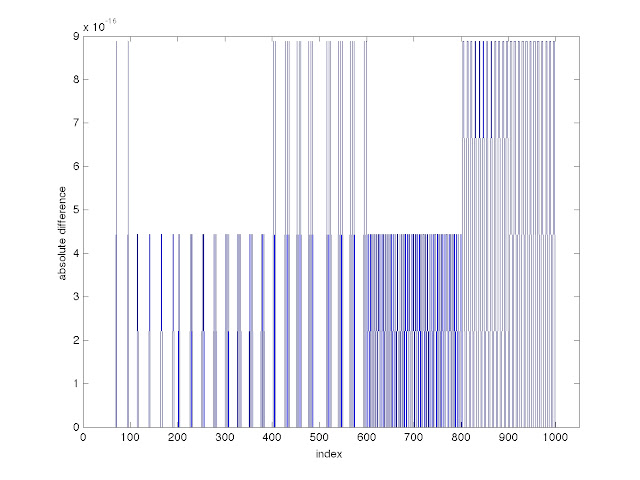 |
The absolute values of the differences between corresponding elements of two arrays, one made with colon notation and the other made with a linspace command.
|
|
When I first made that graph, the horizontal axis went out to 1200. I decided to cut it down, since 1200 is close to number of indices in my X array and I didn't want it to be ambiguous. You can specify a viewing window with the
axis command:
axis([horizontal_min horizontal_max vertical_min vertical_max]).
You'll have to specify the values of horizontal_min, horizontal_max, and their vertical counterparts.
Getting back to making the contour plot, now we need to make the coordinate grid, which I do with the
meshgrid command.
[x,y] = meshgrid(X,Y);
and calculate the function:
z = 6*x.*y - 5*x.^2;
When using arrays, the period before an operator tells MATLAB to go element-by-element. This way, x.*y means x_11*y_11, x_12*y_12, etc. and x.^2 means (x_11)^2, (x_12)^2, and so on.
We're ready to plot now, so let's make a pcolor plot--the default is faceted shading:
What happened? Faceted shading prints the grid, but the grid points are so close together you can't see the function. If you click on the zoom-in button on the top of the figure window and define a viewing window small enough, you can see some of the colorized plot. Typing "axis('auto')" will get you back to the default viewing window, or you can use the zoom-out button. Flat shading comes next, and you can get that by typing "shading('flat')". In general, it will look like a coarse version of what you'll get by typing "shading('interp')" for interpolated shading, but, personally, I can't see a big difference for this example. I'll just show you the interpolated shading.
The axes aren't labeled. You will find fewer things irritate a scientist more than showing them a plot with unlabeled axes. Regardless of what it is you've plotted, or how you're referring to the axes, you label the horizontal and vertical axes, respectively, with the following commands:
xlabel('horizontal_axis_label')
ylabel('vertical_axis_label')
I'm just going to call the axes x and y.
xlabel('x')
ylabel('y')
Let's add a color bar so we have some scale for the function. The command is
colorbar (some things are straightforward).
Right now things should look like this:
If you want to see the plot in grayscale, use the command
colormap. Type in "colormap(gray)" and you'll get the following. You must spell it "gray". If you use the word "grey", it will generate an error.
To get back to the old colormap, type "colormap('default')". If you didn't notice, in the commands the word "default" had single quotes around it, but "gray" didn't. By the way, I got most of the information for grayscale that I wanted with a Google search that pointed me to a webpage made by MATLAB's parent company. Like I said, the help pages aren't always as useful as you'd like them to be. Just in case it wasn't clear, I'm going to switch back to the default colormap.
Now we're ready to draw some contours. If you want to represent different categories with different colors and/or line styles, you should decide what the categories are before proceeding. You could probably change your mind in the middle, but I find that things are generally easier if you know what you want beforehand. I want two categories: contours down to z = -150, which I'll used black, dashed-dotted lines for and contours below z = -150, for which I'll use black, dashed lines. In both cases, I plan on incrementing by 25 and, because I already drew a color bar, I have an idea about the range I need to cover for both categories. The following commands will generate the vectors whose elements indicate where I want contours drawn.
dashdots = -150:25:50;
dashes = -400:25:-175;
Turn the
hold command on by typing "hold on".
You need to assign a handle if you want to label the contours; it doesn't matter what it's called. The handles are taken care of by the things on the left hand side of the equals sign. The right hand side takes care of drawing the contours at the specified levels and the line styles and colors (see "help plot" for line styles and colors). If you know that you don't want to label the contours, you can just type in the commands on the right hand side of the equals sign.
[b,g] = contour(x,y,z, [dashdots], 'k-.');
[c,h] = contour(x,y,z, [dashes], 'k--');
I advise using the semicolons after the commands or MATLAB will echo the values of b, c, g, and h to the screen. After the first command, the dashed-dotted lines closer to the plot center should have appeared and the dashed lines should have shown up after the second command. The plot should look like this:
And now we add labels withe the
clabel command.
clabel(b,g);
clabel(c,h);
And now your plot should look like what I promised:
And now, to save as the highest possible quality jpeg:
print -djpeg labeled_contours
Whatever directory MATLAB is currently working in, that command should leave a file there.
So that does it. If you're using this to make a labeled contour plot, I hope you found it useful. The whole point of writing it the way I did was so that I could reread this post and be able to do it in a few minutes. In the next post I'll start showing some of the plots that can be generated from the SURYA output files.






















